Nanoscale Marvels
From Feynman’s Vision to Bacterial Motors
A vast playroom filled to the brim with nano Lego
In the final days of the 1950s(!) Richard Feynman gave an inspiring lecture at the Annual Physics Society about the future where manipulating individual atoms or even construct nanomachines would be possible. It was titled “There’s Plenty of Room at the Bottom” and each year scientists and engineers are taking steps to realise that vision.

Chip manufacturing capabilities have pushed the boundaries of physics many times over, with modern processors manufactured in a technology that runs individual metal lines 20 nanometers apart. See this transmission electron microscopy image of a metal gate.

These extraordinary capabilities are not limited to manufacturing microprocessors and GPUs. We are beginning to understand precisely the workings of living nanomachines, collections of proteins that form sophisticated engines. And from understanding we’re onto designing structures that have never existed.
How to design a motor and reverse gear from aminoacid chains
I have been drawn to CryoEM structures reveal how the bacterial flagellum rotates and switches direction for its promise of:
discussing movement principles of nanoscale motors
appreciating the details of sophisticated experimental methodologies and modelling the outcomes with tools like ChimeraX
authors augmenting their research with AlphaFold 2
learning about strong hypotheses and detailed analysis of a fascinating engineering solution of bacterial movement.
As I finish reading the paper I can’t help thinking there’s a fractal nature to appreciation of the detail on many of the above points that an expert could elucidate. If you’re one of them please don’t hesitate to lean in. Still I’ll do my best to share the lessons I’ve learned.
I’ll talk about similarities between a DC motor and a bacterial flagellum movement. I will discuss the protein-based structure and how nature has solved the problem of switching from clockwise to counterclockwise rotation at nanoscale. I’ll describe how changes to alignment of individual protein domains lead to creation of various binding pockets. I’ll talk about experimental cryoEM results and how an imperfect alignment of numbers of protein-based staves in neighbouring parts of the engine leads to a desirable wobbling of the engine. I’ll talk about how neighbouring copies of the same protein link up together to create a circular structure.
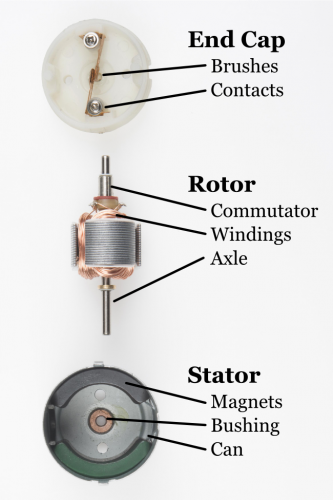
Authors of the paper make several references to terminology of a motor when describing the rotation of a bacterial flagellum.
Stator - which in a DC motor is the source of the constant magnetic field and in the paper is a MotA-MotB protein complex that anchors to the peptidoglycan layer of the bacterial cell wall and forms a proton channel providing source of an electric current.
Bushings - Two sets of rings (Peptidoglycan and Lipid) that stabilise the torque transmission.
Switch - the centre of attention of the article, which through conformational changes of a vast (6 megadaltons) multiprotein complex is able to move from clockwise to counterclockwise rotation and affect the direction of bacterial movement.
Staves - another manufacturing term, although not specifically motor related it paints a vivid picture of a structure composed of a number of identical slats that make up a barrel or a cylinder.
The authors make a number of detailed observations about how the physical and chemical process occurs and we will now dive deeper into it.
Properties
Torque
How is the energy provided by a direct current of protons converted into rotational movement? A much older paper elucidates it, by showing electrostatic interactions between wild type and mutated protein electrostatic potential surface. Bacterial flagellar motors rotate, obtaining power from the membrane gradient of protons.
Type of electrostatic interactions between rotor and stator in the bacterial flagellar motor can be inspected in Figure 4 in the paper. Figure shows various mutations (R->E, D->K) that hindered or increased torque transfer.
Stability
Another piece of interesting structural behaviour I picked up on is the concept of domain swapping. At first I thought this means that proteins detach their domains and swap them out(!) though that made little sense. In fact the meaning behind this is that they slot between each other to strengthen the structure of the overall engine.
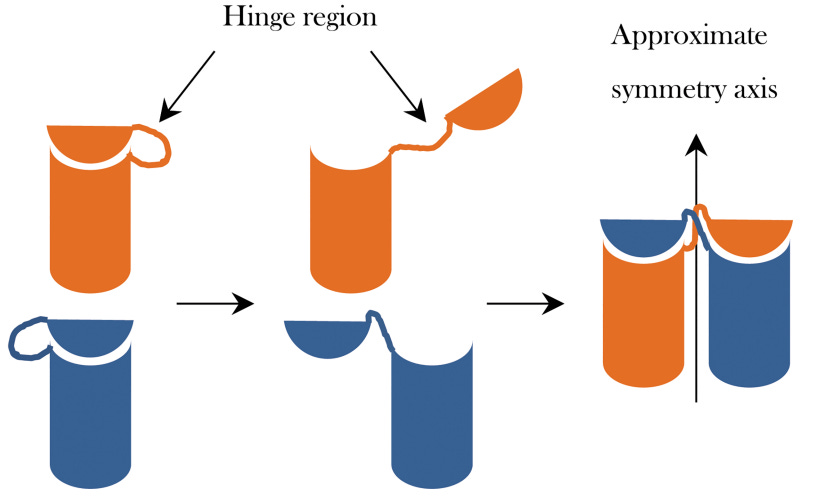
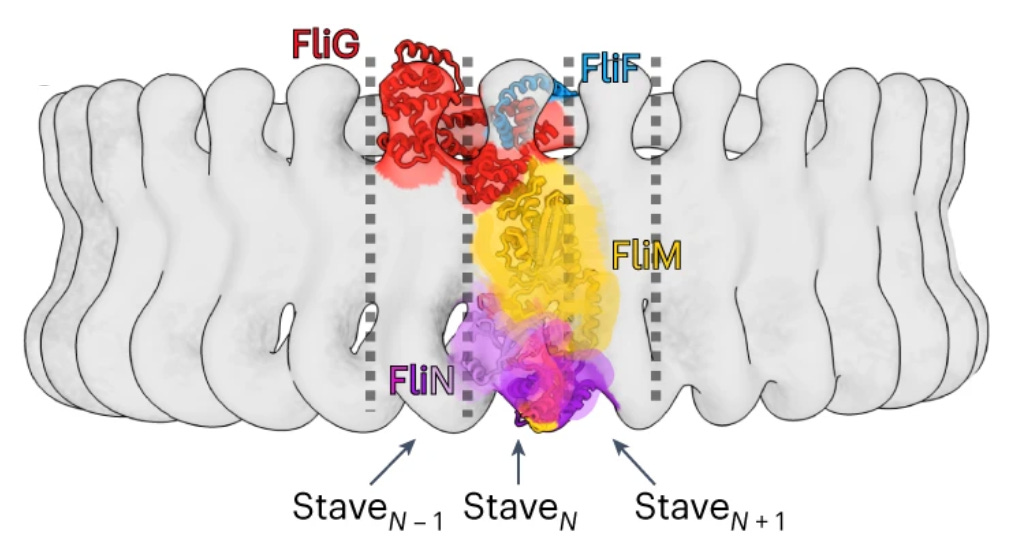
Direction switching
I was blown away by the details of the structural switch mechanism. A single Che-Y bound to a cleft in the FliM part in just one of 30+ staves causes a conformational change where the torque coil (FliG-D5) reorients itself [paper’s Figure 4] and all the other staves follow the reconfiguration like dominoes.
There’s a lot more detail in the paper that I absorbed cursorily, and it leaves me wanting to dig deeper: the impact of a three amino acid deletion mutation PAA on CW/CCW propensity, discussion around the full integer ratio vs other than 3:1 proportion of staves (34 FliM to 102 FliN) of different part of the structure on its low energy minima, perhaps preventing the seizing up of the motor and much more about modelling of the CryoEM measurements. It will have to wait for next time.
Conclusion
We live in extraordinary times. Cutting edge research is available open access and thanks to search engines and AI assistants it’s never been easier to grok the details of an ostensibly dense academic publication. What inspires me most is Prash Singh, the author himself, being incredibly generous with his time to answer a few of my clarifying questions. What an exciting, interconnected world we live in.
#atoms
July 28th addendum
Smarter Every Day just posted a 30 minute video on this topic. It’s a reflection of how many people are seeing the consequences of this research.