Scientific hypotheses directly from data
...can machine learning discover new scientific laws from atom scale imaging?
The scientific approach is driven by the ability to identify patterns and formulate hypotheses that predict future experimental data. At the smallest scales, we employ the language of quantum physics and chemistry. When imaging ensembles of atoms that form materials, the periodic arrangement, including any defects, is of particular interest. Given the emerging capabilities of machine learning, it is reasonable to investigate whether algorithms capable of approximating complex distributions—modern deep neural networks - might potentially uncover deeper relationships within atomic assemblies.
We are beginning to see approaches that utilise machine learning to model interactions and strain within a graphene structure imaged using scanning transmission electron microscopy. This is reflected in Physics and chemistry from parsimonious representations: image analysis via invariant variational autoencoders, published on 2024-08-14. We will explore how imaging studies of spatially periodic materials with atomic-level defects are being transformed by an algorithm developed just a decade ago: Variational Autoencoders.
Before we get to computational materials a quick - equation free - refresher on how VAEs work.
A word to the wise …is sufficient
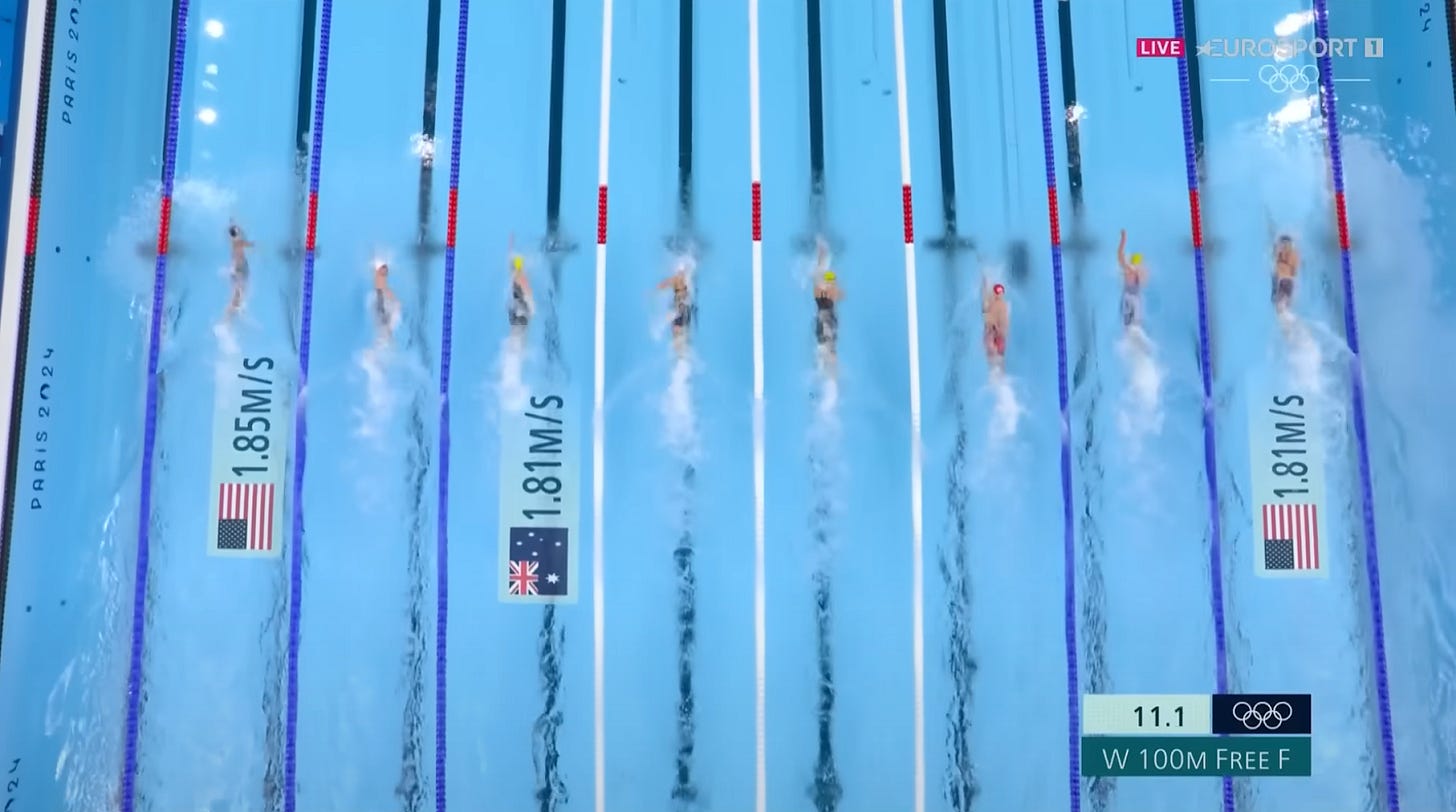
There is something powerful about building systems that can compress or distil a vast amount of information into a small number of data points while still preserving the essence of the story. A recent example from the Olympics demonstrates this. Immense TV audience excitement was generated by observing only three evolving speed values to determine if a swimmer was slowing down or speeding up. The entire complexity of the race was condensed into a few numbers: time and position at the flip turn and distance to leader / speed during the top down view. With a strong mental model of the arena, the swimming pool, the competition, and the prize - an Olympic medal - the audience was able to fully engage in the thrill with no more than ten real-time numbers.
Vivid imagination or veracious reconstruction
We can condense the essence into a few key numbers. But what about a pixel perfect reconstruction of the race? Perhaps swimming, with all its splashes, might be too challenging. But what about Formula 1 racing? With access to all the rigid body metrics of the car: speed, position, steering wheel angle; is it beyond imagination to fully reconstruct the camera view of a race? It can be achieved in computer graphics. Why wouldn’t it be possible to train a neural network to regenerate the image from vast amounts of past examples, given camera parameters and cars data?
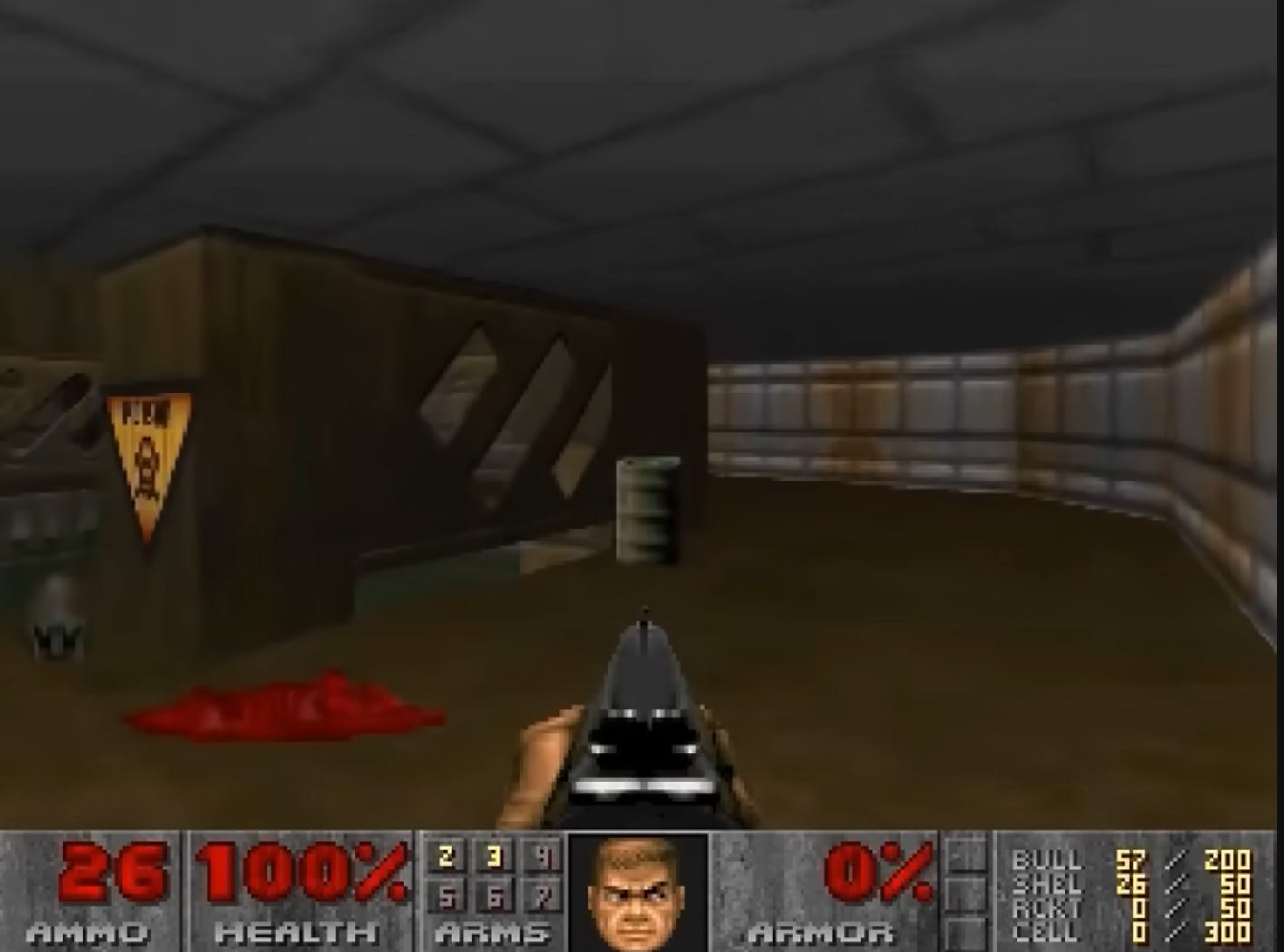
Breaking News: game engine generation. While not as challenging as real time video modelling of a race track with all of its weather and audience variations, Google DeepMind still achieved a mind blowing milestone. On 2024-08-27 it published Diffusion Models Are Real-Time Game Engines GameNGen, modelling the Doom game engine at 20 FPS at fidelity making it - in shortish context time windows - indistinguishable from the real thing for human observers. It shows the power of neural networks ability to deeply understand the entire distribution of input data they have seen during training and capacity for pixel perfect reconstruction.
AE
We discussed distilling a complex situation into a few words: encoding. We also talked about reconstructing a complex scene from just a few coordinates: decoding. This can be achieved by utilising a typical image processing neural architecture: a convolutional neural network, capped with a layer that produces a vector - rather than a typical categorical classification decision; hotdog - not hotdog. Consider this vector as the aforementioned racing car coordinates - a vector of latent space. The reconstruction would "simply" be carried out by an inverse architecture to the one that encodes into latent space. It would take the latent space vector as input, providing the context to reconstruct the original image. What you have there is a non-variational autoencoder.
The V in VAE
What’s the variational trick? Rather than asking the encoder network to predict the vector of length N directly we ask it to predict parameters of an N-dimensional gaussian: the mean and the variance of N dimensions. One can be tempted to whole-hog an NxN covariance matrix but that’s only for special cases.
Now, during training we need to balance two sources of loss
1. Distribution alignment loss
We want the neural network to encode an image in such a way to predict a mean of 0 and variance of 1 for each latent dimension.
2. Reconstruction loss
We want the parameters of the predicted Gaussians used to generate a latent vector to cause the decoder network to regenerate the input faithfully. Meaning we need expressiveness and diversity which we won’t achieve if we always predict the same mean=0 and variance=1.
This tug of war between predicting distribution parameters close to Gaussian normal and infusing the latent space with expressiveness contributes to learning a model that’s expressive, wise - through compression - and generative.
Why go through all this variational trouble?
Adding the distribution component enables the architecture to be generative and produce new, realistic samples. During the variational training, the neural network setup enforces smooth behaviour of the distribution in latent space, ensuring that the samples produced are realistic. The ability to interpolate in low-dimensional latent space between samples seen at the input allows for never-before-seen, but plausible, new structures to be produced.
Let’s talk materials and microscopes
The authors of the paper first test their VAE architectures on toy datasets: images of handwritten digits and suits of cards that underwent rotation and shear, to assess the robustness of the networks in building a latent space that places two images of the same class closer to each other than two images of different classes. After testing the architecture, they apply it to analyse the 'distilled essence' of the meaning of hexagonally arranged carbon atoms with silicon imperfections, as well as collecting atomic microscopy feedback force when it comes into contact with a thin slice of ferroelectric film to determine the walls of individual ferroelectric domains.
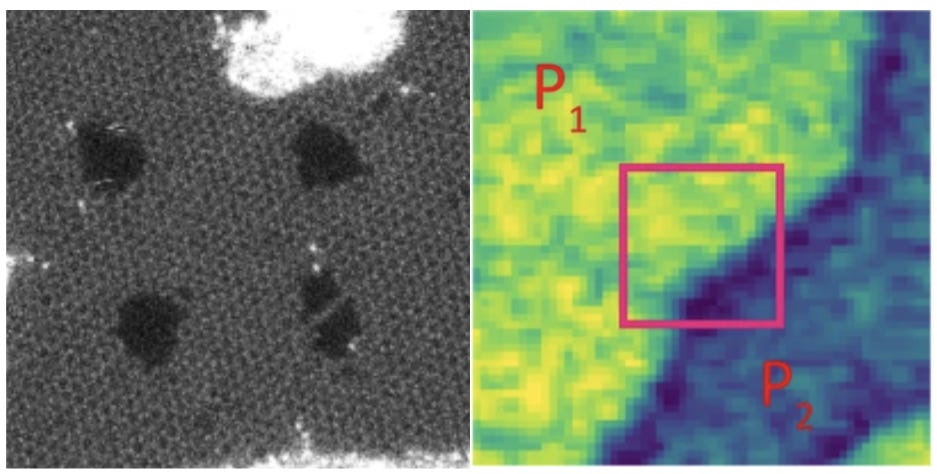
They typically choose very low-dimensional latent spaces. They either pick two dimensions for two latent variables or add a third dimension to introduce an explicit dimension for 2D angular rotation and two dimensions for x and y translation. They also experiment with adding an M-dimensional vector for M classes to be one-hot encoded.
The authors are able to compress the images and derive basic observations about how different elements of the latent space may encode different arrangements of carbon atoms in graphene. They also speculate that a part of the latent space that varies with the image but cannot be easily explained by inspecting them visually may encode strain. I think it’s an early attempt at applying modern ML, and the material scientific conclusions are somewhat underwhelming. That being said, the article serves as a reasonable refresher on VAE architectures.
Future of scientific collaborations
Authors of the paper observe that we live in a world where data, coding researchers and computational power influence each other. As more and more data comes online it is imperative that we move from qualitative inspections to quantitative methods. Especially when inspecting materials where emergent properties not described by typical physics computations is too expensive to be done for every single sample.
And others are not resting. Just this week Google DeepMind published a Science paper on using ML to approximate excited quantum states. But that’s a topic for another time.
#embeddings





