Why patients exhibit resistance to monoclonal antibody therapies?
An investigation of key proteins involved in well regulated human immune system: PD1, IFNG, JAK, STAT and PTEN and how their mutations allow cancer to evade an immune response.
Last week I tried to understand the mechanisms behind a therapy involving antibodies designed to stop tumour cells from deactivating T-cells via the PD-L1 <> PD-1 handshake. There are many such protein pairs, but this one is the most researched and targeted.
Today I want to better understand mechanisms of occasional immune system’s resistance to those therapies, which is discussed in the second half of the paper Cancer immunotherapy with immune checkpoint inhibitors (ICIs): potential, mechanisms of resistance, and strategies for reinvigorating T cell responsiveness when resistance is acquired.
Some patients exhibit acquired resistance, meaning they initially respond to therapy but eventually, after months or sometimes years, the disease comes back. Others never react to treatment in the first place: primary resistance.
Important term: gene pathway—inside a cell, different proteins interact to affect each other's behaviour or trigger the production of more proteins. A pathway can be turned on or off depending on the cell's needs. The “on” signals are often controlled by proteins called kinases, which turn on signals by adding a phosphate group (P) from ATP. The “off” signals are managed by phosphatases, which remove the phosphate group from proteins to stop the signal. Kinase giveth, phosphatase taketh away.
Battling primary resistance
ICIs here are often used in combination with chemotherapies. A more personalised approach may mean looking at a patient's individual biomarkers to see whether and which type of ICI treatment may be successful:
Level of expression of—the perennially interesting—PD-L1 is assessed.
Analysis of the patient's entire exome and screening for the amount of mutations, the so-called tumour mutational burden. The exome is the small 1–2% portion of DNA that encodes actual protein sequences.
Whether there has already been a successful infiltration of the tumour by the patient’s immune system, specifically lymphocytes. This requires a biopsy of the tumour to search for their presence inside the tumour. Interestingly, staining of different immune cell types is achieved via infusing the sample with dyed antibodies designed specifically to bind to them.
We already learned how protein mutations - associated with cancer - found inside the cell can alert the immune system. Proteins inside the cell are routinely chopped up, and their fragments, peptides 8–11 amino acids long, are exposed on the cell surface within a structure resembling a vice or a grip called MHC: major histocompatibility complex. MHC plus peptide is a pMHC. These peptides are sniffed by the immune system.
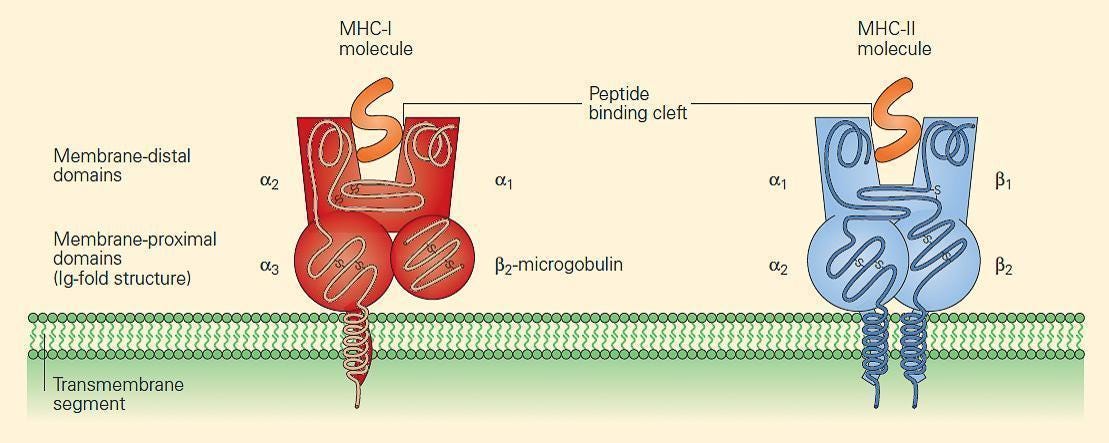
The B2M gene (and therefore its protein) is associated with the stability of MHC. B2M mutations result in non-functioning MHC complexes, which prevent T-cells from triggering an alarm. A review paper cites a melanoma study where patients exhibited these B2M mutations, and that patients with this mutation also did not respond to therapy targeting another therapeutically popular inhibitory receptor CTLA-4.
Important term: epigenetic suppression—silencing of gene expression through environmental (non-mutation) factors. This is reversible and can help the tumour avoid immune detection by reducing the expression of proteins that would otherwise trigger an immune response.
Important term: immunological evasion—tumours can achieve this by down regulating immune-related molecules (like MHC building proteins) or by expressing checkpoint molecules (like PD-L1) that inhibit T-cells action.
Interferon gamma IFNG
Next, let’s better understand the role of IFNG in its default setting and imagine how cancer may cause its dysregulation. IFNG is a small protein (~150 amino acids long) which serves as a signalling molecule produced by many immune cells with the goal of activating and regulating an immune response. It has been found to be involved in a vast number of gene expression pathways.
MHC expression: As mentioned above, the B2M protein is responsible for the stability of the MHC—the tiny vice presenting 8–11 amino acid chains of proteins expressed inside the cell. IFNG stimulates cells to produce more MHC on their surface.
Directly induces cells to increase production of antiviral proteins.
Increases the chance that a mutating cell is killed:
by CD8+ T cells: by stimulating expression of “kill me” signals on the sick cell’s surface
by macrophages, by binding directly to receptors on their surface
IFNG signals via the JAK-STAT pathway (JAK1, JAK2, STAT1 proteins) to build more MHC complexes on the cell surface. If any of those genes undergo a mutation, it disrupts MHC presentation and hence makes cells less visible to the immune system. One can imagine a cell being filled with IFNG, triggering more and more JAK-STAT pathway steps but still no new MHC appearing on the cell surface. Since IFNG is involved in many pathways, it can also lead to upregulation of PD-L1, the protein which “shakes hands” with T-cell’s PD-1, deactivating its attack. Finally, the overabundance of IFNG will lead to T-cell exhaustion, where T-cells don't know what to attack anymore.
Tumour suppressor PTEN
Earlier I talked about kinases and phosphatases as “on” and “off” switches for gene pathways. PTEN is heavily involved in modulating and depressing the cell division rate. When cells need to stop multiplying, PTEN is the one that puts on the brakes. It is also involved in DNA repair by protecting cells from “replication stress”. This replication stress, making inaccurate copies, is of course a source of cancerous mutations. The paper references patients whose genomic profiling found deleterious PTEN mutations coincided with acquired resistance to a PD-1 inhibitor therapy.
Are we there yet?
We’re not there yet with understanding, control and interventions in gene pathways for specific tumours. Attacking something as omnipresent as PD-1 cannot possibly be the limit of our capabilities as researchers and doctors.
Improved analytical tools and experimental techniques can provide clearer insights, leading to better predictive models and personalised therapies. By shining a more informative light on these problems, we move closer to overcoming resistance mechanisms and enhancing patient outcomes in the fight against cancer.
#atoms
Next time we’re looking at something even more complex than immunology. Continual improvement of ways of working for tech teams.



